Oversized and understudied: Researchers begin to uncover the mysterious lives of jumbo bacteriophages

Viruses are tiny but mighty intruders that can be found in the environment including in human bodies, and they can play a variety of roles in ecosystems. Viruses also come in a range of sizes. Some are even larger than bacteria, which scientists thought were rare cases until recently.
Among these large viruses are bacterial viruses called jumbo bacteriophages. Jumbo phages were discovered decades ago, but they have, rather paradoxically, mostly escaped further investigation because scientists were looking on too small of a scale.
Alaina Weinheimer, a Ph.D. candidate, and Frank Aylward, an assistant professor of biological sciences in Virginia Tech’s College of Science and an affiliate faculty member in the Center for Emerging, Zoonotic, Arthropod-borne Pathogens within the Fralin Life Sciences Institute, are seeking to answer a few big questions about these jumbo viruses. In a recent study, Weinheimer constructed and analyzed the genomes of marine jumbo phages, which led to a number of findings about their evolution and ecology.
“We are only beginning to appreciate the diversity and scope of jumbo phages in the environment” said Weinheimer, and the first author on the paper. “We don't quite understand the impacts of jumbo phages yet, and we're just beginning to see that they are quite widespread. This study shows that they are found all throughout the ocean, and they don't necessarily all infect the same type of bacteria.”
Their findings were published in The ISME Journal: Multidisciplinary Journal of Microbial Ecology.
Bacteria are the driving force behind the ocean's nutrient cycles. Despite their position at the bottom of the food chain, bacteria perform vital functions such as photosynthesis and nitrogen fixation, which contribute greatly to the health of marine ecosystems.
When jumbo bacteriophages infect and kill bacteria, they first seize control of the bacteria’s metabolism, transforming the bacteria’s principal functions — such as photosynthesis and transcription — into the replication of more viruses. To release new viruses, the bacteria cell bursts, which also releases the cell’s nutrients and organic material into the sea. These infections shape nutrient cycles in the ocean.
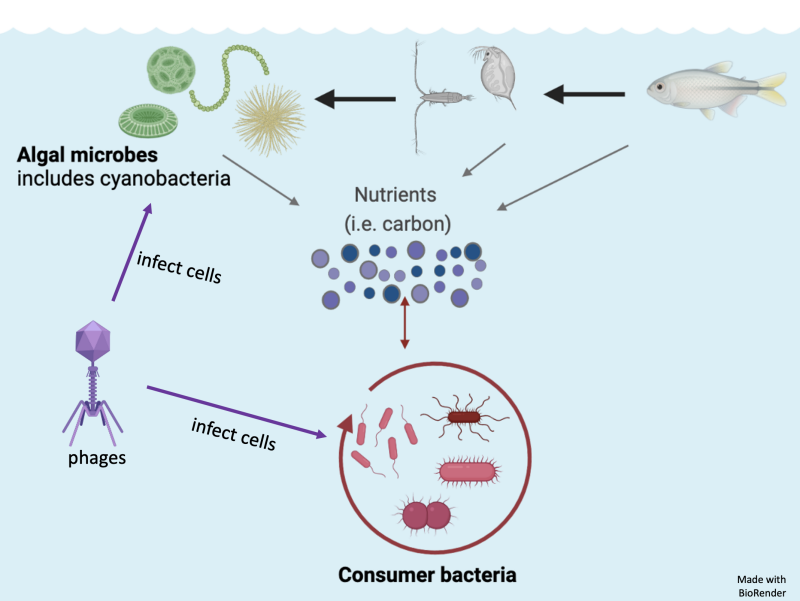
For that reason, researchers are determined to understand how phages change the composition of marine microbial communities and nutrient cycles that bacteria affect. But to study viruses such as these, researchers need to work beyond a basic microscopic level.
Viruses can be incredibly difficult to grow in a lab, so to analyze them, researchers often go out and collect DNA samples from the environment called metagenomes. Each metagenomic sample is composed of DNA from many organisms or entities, which means researchers have to isolate and piece together the genomes of viruses themselves.
From there, the process is like constructing an incomplete puzzle.
“When we sequence DNA in the environment, we first have to break the DNA up into itty bitty pieces,” said Weinheimer. “After they are sequenced, we put the pieces back together into longer stretches of DNA that we think belong to the same genome.”
Because of their particularly complex evolution and genomes, researchers must come up with intricate ways to detect jumbo phages and reconstruct their genomes as accurately and completely as possible.
After some trial and error, Weinheimer and Aylward developed a method that will help other researchers better identify and group jumbo bacteriophages in metagenomes.
With this approach, Weinheimer and Aylward were able to successfully recover 85 high-quality jumbo phage genomes present in the ocean. They then grouped these jumbo phages based on gene content with other known jumbo phages and were able to conclude that certain groups of jumbo phages are more prevalent in surface waters than deeper waters and vice versa.
“Traditional methods typically only look at a fragment of jumbo phage genomes,” said Weinheimer. “But with our approach, we are getting closer to full genomes, and so we're able to better our understanding of the diversity and biology of these phages.”
In their study, Weinheimer and Aylward also suggest that bacteriophages have different routes of evolving such big genomes. While some phages acquire photosynthetic genes to aid in infection efficiency, others will pick up genes that are more vital for combating their host's defenses.
This finding backs up the hypothesis that a defense system arms race is part of the reason behind the impressive size and variety of bacteriophage genomes.
But there is more to be done to understand how complexity emerges or evolves in the virus world and how they evolve with their hosts. Now that Weinheimer and Aylward know where these jumbo phages are more prevalent, they can collect samples to potentially even begin to grow them in the lab.
“By targeting and isolating jumbo phages or growing them in the lab, we may better understand their biology,” said Weinheimer. “A lot of jumbo phages have genes with unknown functions, and we are excited to see what we will find.”
Additionally, their methods can be applied to metagenome samples from other environments such as soils and lakes to begin to see what roles jumbo phages may have in these ecosystems.




